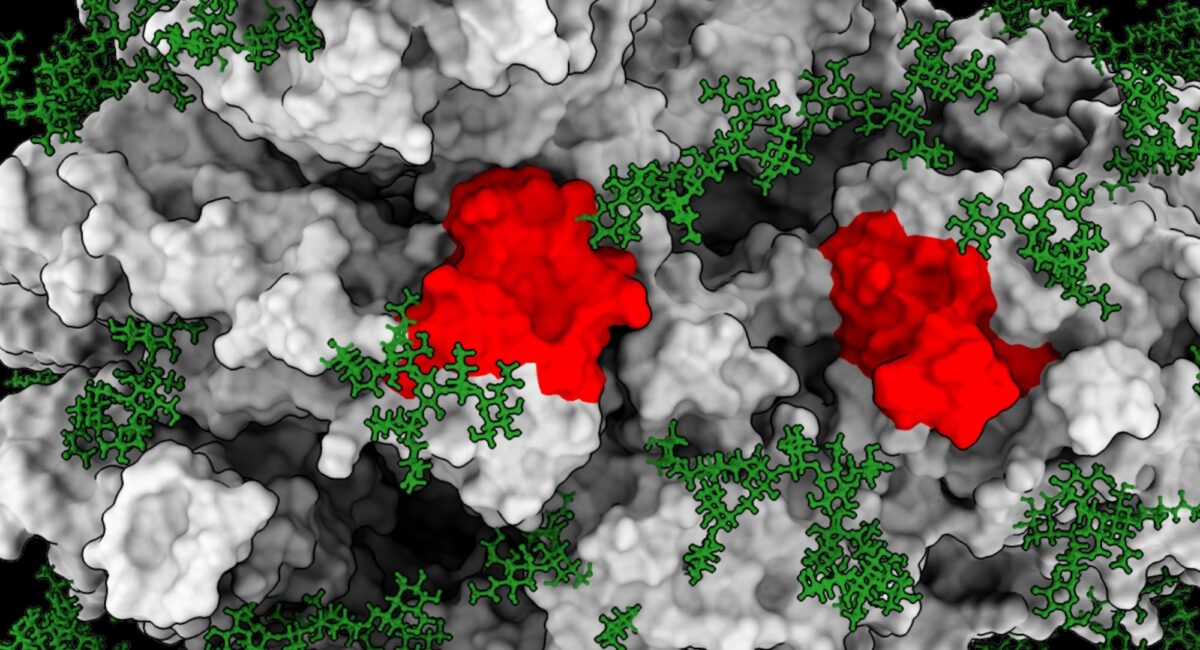

Protein recognition plays a key role in the generation of an inflammatory and an immune response and it is therefore critical to identify the relevant “hot spots” within the protein that trigger the body’s immune defence.
Existing methods and tools attempt the identification of immunogenic regions within proteins, with limited understanding of the potential peptide stability and/or conformation. An innovative approach developed by Professor Gilbert and his team including Dr Patrick Kearns, has been developed for the rapid identification of stable peptides that retain the structural, conformational and/or antigenic properties of a particular region of a full protein.
This new approach is the result of a collaboration with Dr Joseph Marsh, who developed the computational methods which have been patented together with the specific peptide sequences. The computational method were based on rigorous thermodynamic predictions which allowed the identification and prediction of peptide sequences that retain the structural stability and conformational features which characterise that region of the full protein.
An important point to make regarding the context of the rapid progress that has been made in computational protein structure prediction (e.g. Alphafold, ESMfold), is that this unique epitope discovery tool allows new structural models to be available for many proteins, including those that haven't been extensively studied, which extends the possible applications for this method.
The technique can be used to derive short, immunogenic peptides that have potential use in diagnostic and prognostic methods, antibody detection and profiling (‘fingerprinting’), vaccine development and/or monitoring of vaccine performance, and accurate variant detection and measurement. The technology can also be used to develop or identify therapies (e.g. monoclonal antibodies) as well as the application for vaccines.
The team is currently investigating the mechanism of the immune response and inflammatory response to membrane proteins on v SARS/CoV-2.
This technology to identify structurally stable peptides is available for licensing and we are actively seeking partners to work together on co-development or collaboration or to license the new technique and to take it forward into specific applications.
Find out how you can work with the University’s world-class inflammation expertise and how to license this technology or collaborate on further research into the identification and prediction of peptide sequences.
Edinburgh Innovations: Identifying structurally stable peptides
medRxiv: Structural epitope profiling identifies antibodies associated with critical COVID-19 and long COVID

We make it easy to access the University of Edinburgh's multi-disciplinary expertise by matching your needs to the latest research, new technologies and world-class facilities.